Ulrich Lab
Research Publications Group Members Biography Team Leader - Reuter Team Leader - WollscheidThe nuclear actin cytoskeleton in genome stability
Our research focuses on the poorly described and controversial functions of the actin cytoskeleton inside the nucleus, particularly in the context of genome stability.
Myosin VI
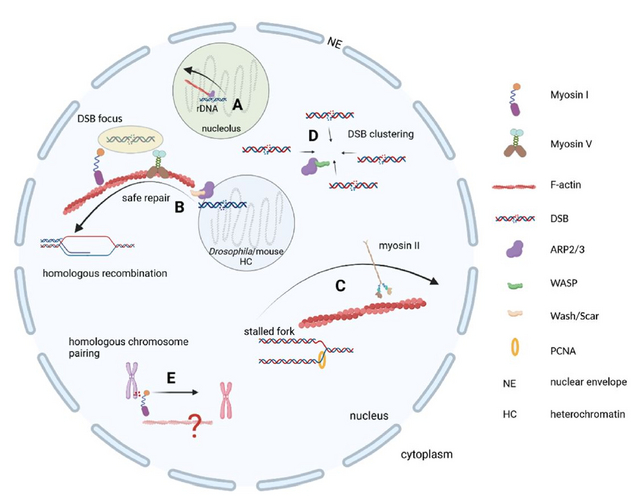
The existence of nuclear filamentous actin (F-actin) has been debated for many decades and is still controversial. However, accumulating evidence from studies in fruit flies as well as mouse and human tissue culture experiments convincingly demonstrate the functional implications of F-actin and associated cytoskeletal proteins in key genome maintenance pathways such as DNA repair (Figure 1) and the response to DNA replication stress.
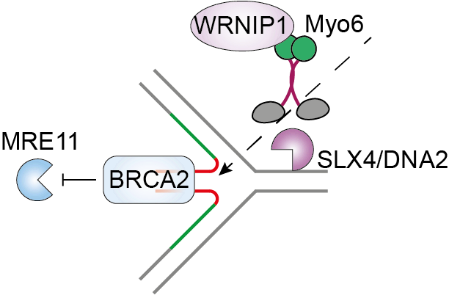
Myosin VI is a unique member of the myosin superfamily as it is the only myosin that transports cargo from the minus to the plus end of actin filaments. In addition to its diverse cytoplasmic functions, myosin VI was described as one of the few myosins with nuclear functions, mainly in the context of transcription. Our current research focuses on the as-yet poorly characterised nuclear functions of myosin VI, with a strong emphasis on genome stability pathways. Recently, we discovered a ubiquitin- and actin-binding dependent function of myosin VI in protecting reversed replication forks from nucleolytic degradation (Shi et al., Nat Commun 2023).
Vision
Conceptually, we follow the idea of a highly dynamic nuclear actin cytoskeleton which, in analogy to its cytoplasmic functions, plays crucial functions in maintaining genome integrity by providing mechanical force for nuclear shape/stability and facilitating mobility and transport within the nuclear compartment. With a repertoire of myosin VI-related tools (DARPins, cell lines, recombinant proteins, etc.), we explore and mechanistically characterise diverse functions of myosin VI in DNA replication and DNA repair to better understand F-actin-related nuclear functions. In the next step, we will expand our analyses to other nuclear cytoskeletal proteins (regulators of actin polymerization, actin-bundling proteins, actin-binding proteins), where we use a wide repertoire of standard molecular biology methods as well as state-of-the-art techniques (e.g. laser micro-irradiation, iPOND, lentiviral transduction, live cell microscopy, DNA fiber assays, etc.).
Recruiting
We are always looking for highly motivated students. If you would like to join our team, please contact us via email.
Hans-Peter Wollscheid
Positions held
- Since 2024:
Independent team leader, Institute of Molecular Biology (IMB), Mainz - 2023 - 2024:
Project leader, University Medical Center (UMC), Mainz - 2018 - 2023:
Independent team leader, Institute of Molecular Biology (IMB), Mainz - 2014 - 2018:
Postdoc, Institute of Molecular Biology (IMB), Mainz - 2009 - 2014:
Postdoc, FIRC Institute of Molecular Oncology (IFOM), Milan
Education
- 2009:
PhD in Biology, University of Konstanz - 2004:
Diploma in Biology, University of Konstanz
Selected publications
Shi J, Hauschulte K, Mikicic I, Maharjan S, Arz V, Strauch T, Heidelberger JB, Schaefer JV, Dreier B, Plückthun A, Beli P, Ulrich HD# and Wollscheid HP# (2023) Nuclear myosin VI maintains replication fork stability. Nat Commun, 14:3787 (#indicates joint correspondence) Link
Wollscheid HP# and Ulrich HD# (2023) Chromatin meets the cytoskeleton: the importance of nuclear actin dynamics and associated motors for genome stability. DNA Repair (Amst), 31:103571 (#indicates joint correspondence) Link
He F*, Wollscheid HP*, Nowicka U, Biancospino M, Valentini E, Ehlinger A, Acconcia F, Magistrati E, Polo S and Walters KJ (2016) Myosin VI contains a compact structural motif that binds to ubiquitin chains. Cell Rep, 14:2683-94 (*indicates joint authorship) Link
Wollscheid HP*, Biancospino M*, He F*, Magistrati E, Molteni E, Lupia M, Soffientini P, Rottner K, Cavallaro U, Pozzoli U, Mapelli M, Walters KJ and Polo S (2016) Diverse functions of myosin VI elucidated by an isoform-specific α-helix domain. Nat Struct Mol Biol, 23:300-8 (*indicates joint authorship) Link
